 |
The amount of code in the world is exploding, as is the amount of code in any given program. Today, essentially all application programs take advantage of prewritten libraries of code--at the very least the runtime library of the chosen programming language(s), and usually many other existing components as well, for graphical interfaces, database access, parsing data formats, and so forth. The analogy to natural genetic recombination is quite strong: Computer source code as genome; the software build process as embryological development; the resulting executable binary as phenotype. The unit of selection is generally at the phenotypic level, or sometimes at the level an entire operating system/applications environment.
A main place where the analogy breaks down is that in manufactured computers, but not in the natural world, there are two distinct routes to producing a phenotype. The extreme `copy anything' ability of digital computers means that source code is not required for to produce a duplicate of a phenotype. Source code is a requirement, in practical terms, for significant evolution via mutation and recombination.
Commercial software is traditionally distributed by direct copying of precompiled binary programs while guarding access to the `germ line' source code, largely to ensure that nobody else has the ability to evolve the line. In that context, the rapidly-growing corpus of `open source' software is of particular interest. With source code always available and reusable by virtue of the free software licensing terms, an environment supporting much more rapid evolution is created. The traditional closed-source `protect the germ line at all cost' model is reminiscent of, say, mammalian evolution; by contrast the free software movement is more like anything-goes bacterial evolution, with the possibility of acquiring code from the surrounding environment and in any event displaying a surprising range of `gene mobility', as when genes for antibiotic drug resistance jump between species. There is therefore reason to expect open source code, on average, to evolve at a faster rate than closed source, at least up to some level of complexity depending on design where the chances of new code being useful rather than disruptive become negligible.
As software systems grow, and software components swallow each other and are in turn swallowed, and older `legacy systems' are wrapped with new interface layers and kept in place, we are arriving at the situation where actually reading fragments of source code tells us less and less about how--if at all--that code ever affects the aggregate system behavior. As this trend accelerates, tools and techniques from biological analysis are likely to be increasingly useful.
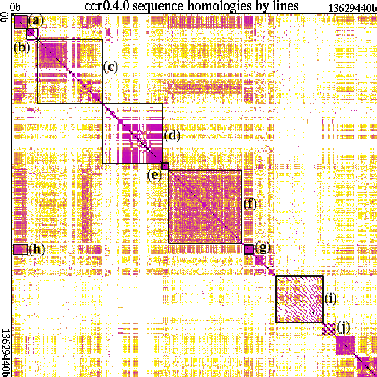
Figure 3 presents another view of the ccr genome, using the `dotplot' program [Helfman, 1996] for visualizing large data sets. This view shows the ccr genome plotted against itself, using lines of code as the fundamental unit of similarity; there is a black line representing perfect overlap down the main diagonal. Dark areas significantly off the main diagonal represent similarities between widely-separated code regions; squares on the diagonal represent `cohesive' regions with more similarity within than without. `Looking under the hood', we find that often such regions either are or are components of larger functional units--`genes'--within the genome. Several such genes have been highlighted with black outlines: Region (a) codes for ccr's web server/client program; (b) is the configuration system that guides the overall system ontogeny; regions (c)-(e) are separately-evolved code segments (for JPEG images, long integer manipulation, and regular expressions, respectively) that have become incorporated, essentially unchanged, into ccr. The large region near the middle of the genome (f) contains the ccr core components themselves. Region (g) deals with processing animated images; interestingly, it shows a perceptible overlap (h) with the web code (a) even though their functions are very different and in fact they are expressed in different languages (C++ (a) vs C (g))--but, it turns out, both were created by the same programmer. Semi-automated project historical notes (i) display a distinctive pattern, as does program-generated Postscript documentation (j).
Figure 4 compares ccr to a different `species'--the `GNU Image Manipulation Program' (GIMP) [Mattis and Kimball, 2000]. There are fewer dark regions, reflecting a generally lower degree of similarity between the code sequences. The dark region (a) is aligned with Figure 3(b) and reveals that both systems use evolved variants of the same `autoconf' developmental control system--though the region is rectangular indicating that GIMP's instantiation of the code is bigger than ccr's. A large `internationalization' segment (b)--allowing GIMP to operate in some eleven natural languages--is strikingly different than almost everything in ccr.
A surprising element in this comparison are two short lines (c)--black diagonals indicating identical sequences. The relevant sequence is the GNU regular expression package, which is used in both ccr (Figure 3(e)) and the GIMP, and is a good example of a highly useful gene incorporated into multiple different applications out of the free software environment. Two short black lines, aligned vertically, show that the GIMP contains two identical copies of the GNU regex gene. Rather than being wasteful--as traditional software practices might have construed it--such gene duplication reduces epistasis and increases evolvability; one copy is deep in the application core and the other in a relatively peripheral `plug-in scripting' segment. Furthermore, from one point of view it's odd that this gene appears at all, because regular expression support is actually a required part of any POSIX-conforming operating system. Yet, like complex living systems, both ccr and the GIMP acquired captive regex `organelles' instead, reducing their vulnerability to environmental variations and increasing their ecological range.